Use of eFEL on the models downloaded from the Neocortical Microcircuit Portal¶
Requirements: - Python 3.9+, including Pip (https://pip.readthedocs.org) - A version of Neuron (with Python support) installed on your computer (for instruction, see https://bbp.epfl.ch/nmc-portal/tools)
Make matplotlib plots show up in the notebook:
%matplotlib inline
Install the eFeature Extraction Library:
!pip install efel
import efel
Get a model package from the website
!curl -o L5_TTPC2.zip https://bbp.epfl.ch/nmc-portal/assets/documents/static/downloads-zip/L5_TTPC2_cADpyr232_1.zip
Unzip the model package:
!unzip L5_TTPC2.zip
Change directory to the model package directory:
import os
os.chdir('L5_TTPC2_cADpyr232_1')
Compile the Neuron mechanisms (if this fails, you might not have installed Neuron correctly)
!nrnivmodl ./mechanisms
Import the model package in Python, and run it:
import run
run.main(plot_traces=True)
Warning: no DISPLAY environment variable.
--No graphics will be displayed.
Loading constants
Setting temperature to 34.000000 C
Setting simulation time step to 0.025000 ms
1
1
1
Loading cell cADpyr232_L5_TTPC2_8052133265
Attaching stimulus electrodes
Setting up step current clamp: amp=0.593063 nA, delay=700.000000 ms, duration=2000.000000 ms
Setting up hypamp current clamp: amp=-0.286011 nA, delay=0.000000 ms, duration=3000.000000 ms
Attaching recording electrodes
Setting simulation time to 3s for the step currents
Disabling variable timestep integration
Running for 3000.000000 ms
Soma voltage for step 1 saved to: python_recordings/soma_voltage_step1.dat
Loading cell cADpyr232_L5_TTPC2_8052133265
Attaching stimulus electrodes
Setting up step current clamp: amp=0.642485 nA, delay=700.000000 ms, duration=2000.000000 ms
Setting up hypamp current clamp: amp=-0.286011 nA, delay=0.000000 ms, duration=3000.000000 ms
Attaching recording electrodes
Setting simulation time to 3s for the step currents
Disabling variable timestep integration
Running for 3000.000000 ms
Soma voltage for step 2 saved to: python_recordings/soma_voltage_step2.dat
Loading cell cADpyr232_L5_TTPC2_8052133265
Attaching stimulus electrodes
Setting up step current clamp: amp=0.691907 nA, delay=700.000000 ms, duration=2000.000000 ms
Setting up hypamp current clamp: amp=-0.286011 nA, delay=0.000000 ms, duration=3000.000000 ms
Attaching recording electrodes
Setting simulation time to 3s for the step currents
Disabling variable timestep integration
Running for 3000.000000 ms
Soma voltage for step 3 saved to: python_recordings/soma_voltage_step3.dat
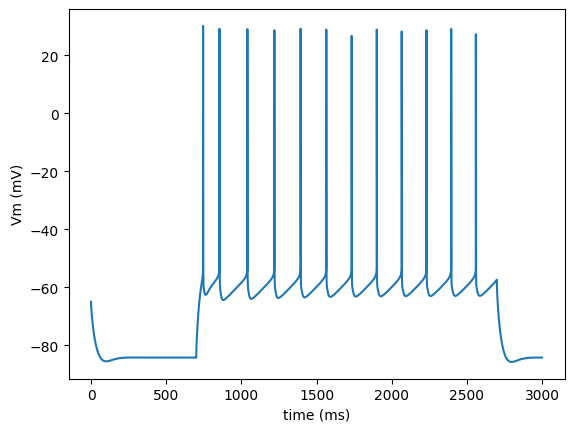
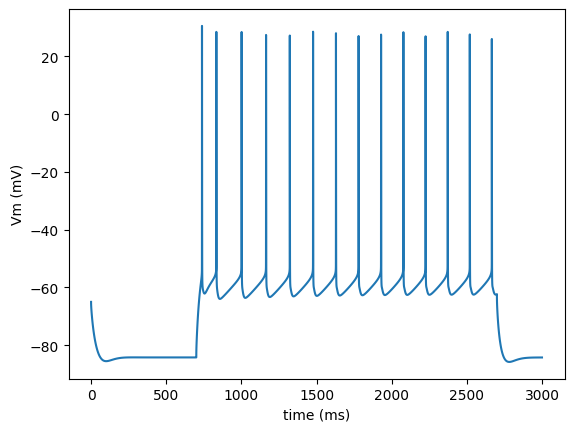
Load the output of the model package in numpy array
import numpy
times = []
voltages = []
for step_number in range(1,4):
data = numpy.loadtxt('python_recordings/soma_voltage_step%d.dat' % step_number)
times.append(data[:, 0])
voltages.append(data[:, 1])
Prepare the traces for the eFEL
traces = []
for step_number in range(3):
trace = {}
trace['T'] = times[step_number]
trace['V'] = voltages[step_number]
trace['stim_start'] = [700]
trace['stim_end'] = [2700]
traces.append(trace)
Run the eFEL on the trace
feature_values = efel.get_feature_values(traces, ['mean_frequency', 'adaptation_index2', 'ISI_CV', 'doublet_ISI', 'time_to_first_spike', 'AP_height', 'AHP_depth_abs', 'AHP_depth_abs_slow', 'AHP_slow_time', 'AP_width', 'peak_time', 'AHP_time_from_peak'])
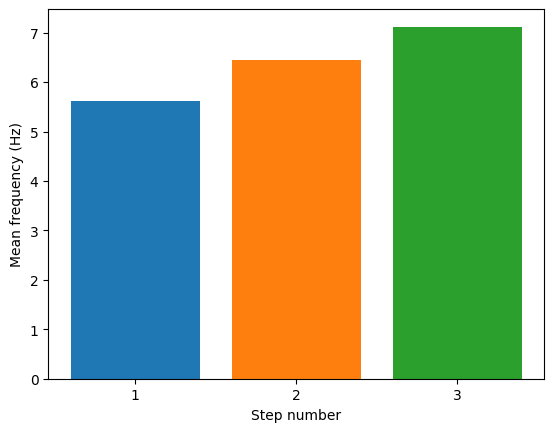
Plot frequencies over steps
import pylab
for step_number in range(3):
pylab.bar(step_number, feature_values[step_number]['mean_frequency'][0], align='center')
pylab.ylabel('Mean frequency (Hz)')
pylab.xlabel('Step number')
pylab.xticks(range(3), range(1,4))
pylab.show()
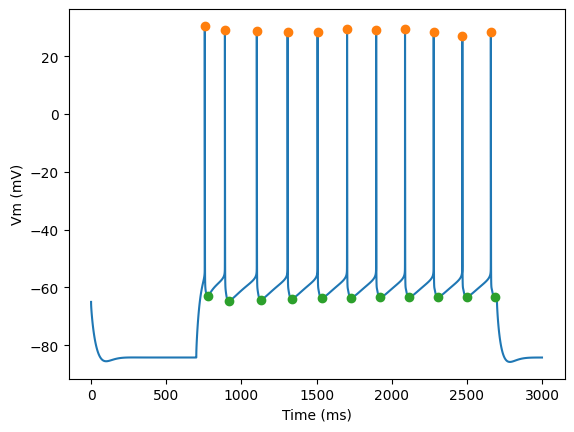
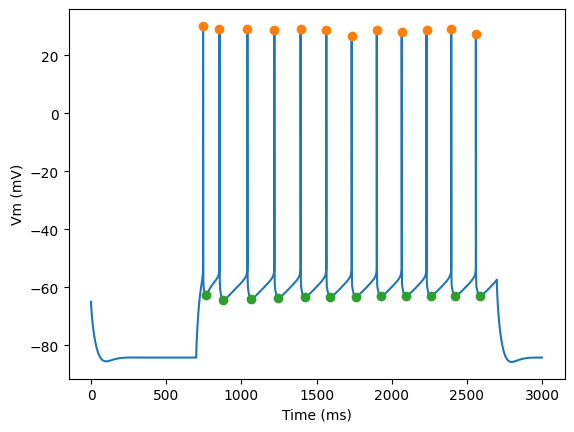
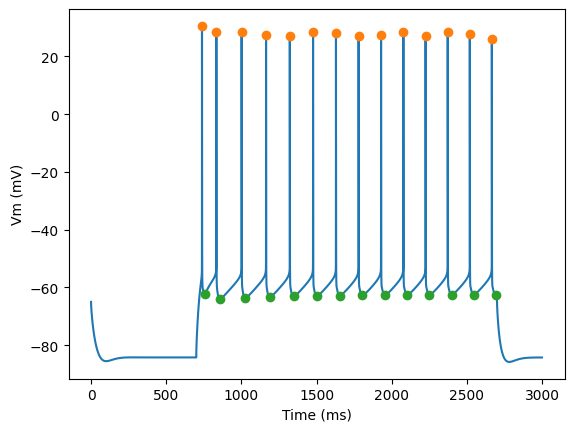
Plot AP height and AHP depth
for step_number in range(3):
time = times[step_number]
voltage = voltages[step_number]
peak_times = feature_values[step_number]['peak_time']
ahp_time = feature_values[step_number]['AHP_time_from_peak']
ap_heights = feature_values[step_number]['AP_height']
AHP_depth_abss = feature_values[step_number]['AHP_depth_abs']
pylab.plot(time,voltage)
pylab.plot(peak_times, ap_heights, 'o')
pylab.plot(peak_times+ahp_time, AHP_depth_abss, 'o')
pylab.xlabel('Time (ms)')
pylab.ylabel('Vm (mV)')
pylab.show()