eFeature descriptions
A pdf document describing the eFeatures is available here.
Not every eFeature has a description in this document yet, the complete set will be available shortly.
Implemented eFeatures (to be continued)
Spike event features
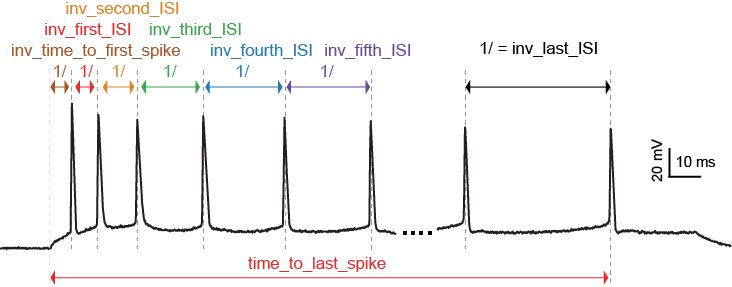
LibV1 : time_to_first_spike
Time from the start of the stimulus to the maximum of the first peak
Required features: peak_time
Units: ms
Pseudocode:
time_to_first_spike = peaktime[0] - stimstart
LibV5 : time_to_second_spike
Time from the start of the stimulus to the maximum of the second peak
Required features: peak_time
Units: ms
Pseudocode:
time_to_second_spike = peaktime[1] - stimstart
LibV5 : inv_time_to_first_spike
1.0 over time to first spike (times 1000 to convert it to Hz); returns 0 when no spike
Required features: time_to_first_spike
Units: Hz
Pseudocode:
if len(time_to_first_spike) > 0: inv_time_to_first_spike = 1000.0 / time_to_first_spike[0] else: inv_time_to_first_spike = 0
Python efeature : ISI_values
The interspike intervals (i.e. time intervals) between adjacent peaks.
Required features: peak_time (ms)
Units: ms
Pseudocode:
isi_values = numpy.diff(peak_time)[1:]
LibV1 : doublet_ISI
The time interval between the first two peaks
Required features: peak_time (ms)
Units: ms
Pseudocode:
doublet_ISI = peak_time[1] - peak_time[0]
LibV5 : all_ISI_values
The interspike intervals, i.e., the time intervals between adjacent peaks.
Required features: peak_time (ms)
Units: ms
Pseudocode:
all_isi_values_vec = numpy.diff(peak_time)
Python efeature : inv_first_ISI, inv_second_ISI, inv_third_ISI, inv_fourth_ISI, inv_fifth_ISI, inv_last_ISI
1.0 over first/second/third/fourth/fith/last ISI; returns 0 when no ISI
Required features: peak_time (ms)
Units: Hz
Pseudocode:
all_isi_values_vec = numpy.diff(peak_time) if len(all_isi_values_vec) > 0: inv_first_ISI = 1000.0 / all_isi_values_vec[0] else: inv_first_ISI = 0 if len(all_isi_values_vec) > 1: inv_second_ISI = 1000.0 / all_isi_values_vec[1] else: inv_second_ISI = 0 if len(all_isi_values_vec) > 2: inv_third_ISI = 1000.0 / all_isi_values_vec[2] else: inv_third_ISI = 0 if len(all_isi_values_vec) > 3: inv_fourth_ISI = 1000.0 / all_isi_values_vec[3] else: inv_fourth_ISI = 0 if len(all_isi_values_vec) > 4: inv_fifth_ISI = 1000.0 / all_isi_values_vec[4] else: inv_fifth_ISI = 0 if len(all_isi_values_vec) > 0: inv_last_ISI = 1000.0 / all_isi_values_vec[-1] else: inv_last_ISI = 0
Python efeature: inv_ISI_values
Computes all inverse spike interval values.
Required features: peak_time (ms)
Units: Hz
Pseudocode:
all_isi_values_vec = numpy.diff(peak_time) inv_isi_values = 1000.0 / all_isi_values_vec
LibV5 : time_to_last_spike
time from stimulus start to last spike
Required features: peak_time (ms), stimstart (ms)
Units: ms
Pseudocode:
if len(peak_time) > 0: time_to_last_spike = peak_time[-1] - stimstart else: time_to_last_spike = 0
Python efeature : spike_count
number of spikes in the trace, including outside of stimulus interval
Required features: LibV1:peak_indices
Units: constant
Pseudocode:
spike_count = len(peak_indices)
Note: “spike_count” is the new name for the feature “Spikecount”. “Spikecount”, while still available, will be removed in the future.
Python efeature : spike_count_stimint
number of spikes inside the stimulus interval
Required features: LibV1:peak_time
Units: constant
Pseudocode:
peaktimes_stimint = numpy.where((peak_time >= stim_start) & (peak_time <= stim_end)) spike_count_stimint = len(peaktimes_stimint)
Note: “spike_count_stimint” is the new name for the feature “Spikecount_stimint”. “Spikecount_stimint”, while still available, will be removed in the future.
LibV5 : number_initial_spikes
number of spikes at the beginning of the stimulus
Required features: LibV1:peak_time
Required parameters: initial_perc (default=0.1)
Units: constant
Pseudocode:
initial_length = (stimend - stimstart) * initial_perc number_initial_spikes = len(numpy.where( \ (peak_time >= stimstart) & \ (peak_time <= stimstart + initial_length)))
LibV1 : mean_frequency
The mean frequency of the firing rate
Required features: stim_start, stim_end, LibV1:peak_time
Units: Hz
Pseudocode:
condition = np.all((stim_start < peak_time, peak_time < stim_end), axis=0) spikecount = len(peak_time[condition]) last_spike_time = peak_time[peak_time < stim_end][-1] mean_frequency = 1000 * spikecount / (last_spike_time - stim_start)
LibV5 : ISI_semilog_slope
The slope of a linear fit to a semilog plot of the ISI values.
Attention: the 1st ISI is not taken into account unless ignore_first_ISI is set to 0. See Python efeature: ISIs feature for more details.
Required features: t, V, stim_start, stim_end, ISI_values
Units: ms
Pseudocode:
x = range(1, len(ISI_values)+1) log_ISI_values = numpy.log(ISI_values) slope, _ = numpy.polyfit(x, log_ISI_values, 1) ISI_semilog_slope = slope
Python efeature : ISI_log_slope
The slope of a linear fit to a loglog plot of the ISI values.
Attention: the 1st ISI is not taken into account unless ignore_first_ISI is set to 0. See Python efeature: ISIs feature for more details.
Required features: t, V, stim_start, stim_end, ISI_values
Units: ms
Pseudocode:
log_x = numpy.log(range(1, len(ISI_values)+1)) log_ISI_values = numpy.log(ISI_values) slope, _ = numpy.polyfit(log_x, log_ISI_values, 1) ISI_log_slope = slope
Python efeature : ISI_log_slope_skip
The slope of a linear fit to a loglog plot of the ISI values, but not taking into account the first ISI values.
The proportion of ISI values to be skipped is given by spike_skipf (between 0 and 1). However, if this number of ISI values to skip is higher than max_spike_skip, then max_spike_skip is taken instead.
Required features: t, V, stim_start, stim_end, ISI_values
Parameters: spike_skipf (default=0.1), max_spike_skip (default=2)
Units: ms
Pseudocode:
start_idx = min([max_spike_skip, round((len(ISI_values) + 1) * spike_skipf)]) ISI_values = ISI_values[start_idx:] log_x = numpy.log(range(1, len(ISI_values)+1)) log_ISI_values = numpy.log(ISI_values) slope, _ = numpy.polyfit(log_x, log_ISI_values, 1) ISI_log_slope = slope
Python efeature : ISI_CV
The coefficient of variation of the ISIs.
Attention: the 1st ISI is not taken into account unless ignore_first_ISI is set to 0. See Python efeature: ISIs feature for more details.
Required features: ISIs
Units: constant
Pseudocode:
ISI_mean = numpy.mean(ISI_values) ISI_CV = np.std(isi_values, ddof=1) / ISI_mean
Python efeature : irregularity_index
Mean of the absolute difference of all ISIs, except the first one (see Python efeature: ISIs feature for more details.)
The first ISI can be taken into account if ignore_first_ISI is set to 0.
Required features: ISI_values
Units: ms
Pseudocode:
irregularity_index = numpy.mean(numpy.absolute(ISI_values[1:] - ISI_values[:-1]))
LibV1 : adaptation_index
Normalized average difference of two consecutive ISIs, skipping the first ISIs
The proportion of ISI values to be skipped is given by spike_skipf (between 0 and 1). However, if this number of ISI values to skip is higher than max_spike_skip, then max_spike_skip is taken instead.
The adaptation index is zero for a constant firing rate and bigger than zero for a decreasing firing rate
Required features: stim_start, stim_end, peak_time
Parameters: offset (default=0), spike_skipf (default=0.1), max_spike_skip (default=2)
Units: constant
Pseudocode:
# skip the first ISIs peak_selection = [peak_time >= stim_start - offset, peak_time <= stim_end - offset] spike_time = peak_time[numpy.all(peak_selection, axis=0)] start_idx = min([max_spike_skip, round(len(spike_time) * spike_skipf)]) spike_time = spike_time[start_idx:] # compute the adaptation index ISI_values = spike_time[1:] - spike_time[:-1] ISI_sum = ISI_values[1:] + ISI_values[:-1] ISI_sub = ISI_values[1:] - ISI_values[:-1] adaptation_index = numpy.mean(ISI_sum / ISI_sub)
LibV1 : adaptation_index_2
Normalized average difference of two consecutive ISIs, starting at the second ISI
The adaptation index is zero for a constant firing rate and bigger than zero for a decreasing firing rate
Required features: stim_start, stim_end, peak_time
Parameters: offset (default=0)
Units: constant
Pseudocode:
# skip the first ISI peak_selection = [peak_time >= stim_start - offset, peak_time <= stim_end - offset] spike_time = peak_time[numpy.all(peak_selection, axis=0)] spike_time = spike_time[1:] # compute the adaptation index ISI_values = spike_time[1:] - spike_time[:-1] ISI_sum = ISI_values[1:] + ISI_values[:-1] ISI_sub = ISI_values[1:] - ISI_values[:-1] adaptation_index = numpy.mean(ISI_sum / ISI_sub)
Python efeature : burst_mean_freq
The mean frequency during a burst for each burst
If burst_ISI_indices did not detect any burst beginning, then the spikes are not considered to be part of any burst
Required features: burst_ISI_indices, peak_time
Units: Hz
Pseudocode:
if burst_ISI_indices is None: return None elif len(burst_ISI_indices) == 0: return [] burst_mean_freq = [] burst_index = numpy.insert( burst_index_tmp, burst_index_tmp.size, len(peak_time) - 1 ) # 1st burst span = peak_time[burst_index[0]] - peak_time[0] N_peaks = burst_index[0] + 1 burst_mean_freq.append(N_peaks * 1000 / span) for i, burst_idx in enumerate(burst_index[:-1]): if burst_index[i + 1] - burst_idx != 1: span = peak_time[burst_index[i + 1]] - peak_time[burst_idx + 1] N_peaks = burst_index[i + 1] - burst_idx burst_mean_freq.append(N_peaks * 1000 / span) return burst_mean_freq
LibV5 : strict_burst_mean_freq
The mean frequency during a burst for each burst
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: burst_begin_indices, burst_end_indices, peak_time
Units: Hz
Pseudocode:
if burst_begin_indices is None or burst_end_indices is None: strict_burst_mean_freq = None else: strict_burstmean_freq = ( (burst_end_indices - burst_begin_indices + 1) * 1000 / ( peak_time[burst_end_indices] - peak_time[burst_begin_indices] ) )
Python efeature : burst_number
The number of bursts
Required features: burst_mean_freq
Units: constant
Pseudocode:
burst_number = len(burst_mean_freq)
Python efeature : strict_burst_number
The number of bursts
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: strict_burst_mean_freq
Units: constant
Pseudocode:
burst_number = len(strict_burst_mean_freq)
Python efeature : interburst_voltage
The voltage average in between two bursts
Iterating over the burst ISI indices determine the last peak before the burst. Starting 5 ms after that peak take the voltage average until 5 ms before the first peak of the subsequent burst.
Required features: burst_ISI_indices, peak_indices
Units: mV
Pseudocode:
interburst_voltage = [] for idx in burst_ISI_idxs: ts_idx = peak_idxs[idx] t_start = time[ts_idx] + 5 start_idx = numpy.argwhere(time < t_start)[-1][0] te_idx = peak_idxs[idx + 1] t_end = time[te_idx] - 5 end_idx = numpy.argwhere(time > t_end)[0][0] interburst_voltage.append(numpy.mean(voltage[start_idx:end_idx + 1]))
LibV5 : strict_interburst_voltage
The voltage average in between two bursts
Iterating over the burst indices determine the first peak of each burst. Starting 5 ms after the previous peak, take the voltage average until 5 ms before the peak.
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: burst_begin_indices, peak_indices
Units: mV
Pseudocode:
interburst_voltage = [] for idx in burst_begin_idxs[1:]: ts_idx = peak_idxs[idx - 1] t_start = t[ts_idx] + 5 start_idx = numpy.argwhere(t < t_start)[-1][0] te_idx = peak_idxs[idx] t_end = t[te_idx] - 5 end_idx = numpy.argwhere(t > t_end)[0][0] interburst_voltage.append(numpy.mean(v[start_idx:end_idx + 1]))
LibV5 : interburst_min_values
The minimum voltage between the end of a burst and the next spike.
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: peak_indices, burst_end_indices
Units: mV
Pseudocode:
interburst_min = [ numpy.min( v[peak_indices[i]:peak_indices[i + 1]] ) for i in burst_end_indices if i + 1 < len(peak_indices) ]
LibV5 : postburst_min_values
The minimum voltage after the end of a burst.
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: peak_indices, burst_end_indices
Units: mV
Pseudocode:
interburst_min = [ numpy.min( v[peak_indices[i]:peak_indices[i + 1]] ) for i in burst_end_indices if i + 1 < len(peak_indices) ] if len(postburst_min) < len(burst_end_indices): if t[burst_end_indices[-1]] < stim_end: end_idx = numpy.where(t >= stim_end)[0][0] postburst_min.append(numpy.min( v[peak_indices[burst_end_indices[-1]]:end_idx] )) else: postburst_min.append(numpy.min( v[peak_indices[burst_end_indices[-1]]:] ))
LibV5 : time_to_interburst_min
The time between the last spike of a burst and the minimum between that spike and the next.
This implementation does not assume that every spike belongs to a burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Default value is 2.0.
Required features: peak_indices, burst_end_indices, peak_time
Units: ms
Pseudocode:
time_to_interburst_min = [ t[peak_indices[i] + numpy.argmin( v[peak_indices[i]:peak_indices[i + 1]] )] - peak_time[i] for i in burst_end_indices if i + 1 < len(peak_indices) ]
Python efeature : single_burst_ratio
Length of the second isi over the median of the rest of the isis. The first isi is not taken into account, because it could bias the feature. See LibV1: ISI_values feature for more details.
If ignore_first_ISI is set to 0, then signle burst ratio becomes the length of the first isi over the median of the rest of the isis.
Required features: ISI_values
Units: constant
Pseudocode:
single_burst_ratio = ISI_values[0] / numpy.mean(ISI_values)
Spike shape features
LibV1 : peak_time
The times of the maxima of the peaks
Required features: LibV5:peak_indices
Units: ms
Pseudocode:
peak_time = time[peak_indices]
LibV1 : peak_voltage
The voltages at the maxima of the peaks
Required features: LibV5:peak_indices
Units: mV
Pseudocode:
peak_voltage = voltage[peak_indices]
LibV1 : AP_height
Same as peak_voltage: The voltages at the maxima of the peaks
Required features: LibV1:peak_voltage
Units: mV
Pseudocode:
AP_height = peak_voltage
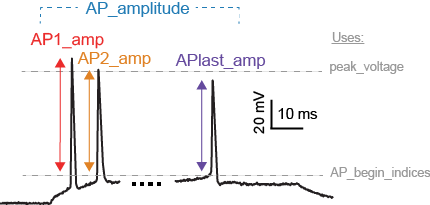
LibV1 : AP_amplitude, AP1_amp, AP2_amp, APlast_amp
The relative height of the action potential from spike onset
Required features: LibV5:AP_begin_indices, LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
AP_amplitude = peak_voltage - voltage[AP_begin_indices] AP1_amp = AP_amplitude[0] AP2_amp = AP_amplitude[1] APlast_amp = AP_amplitude[-1]
LibV5 : mean_AP_amplitude
The mean of all of the action potential amplitudes
Required features: LibV1:AP_amplitude (mV)
Units: mV
Pseudocode:
mean_AP_amplitude = numpy.mean(AP_amplitude)
LibV2 : AP_Amplitude_change
Difference of the amplitudes of the second and the first action potential divided by the amplitude of the first action potential
Required features: LibV1:AP_amplitude
Units: constant
Pseudocode:
AP_amplitude_change = (AP_amplitude[1:] - AP_amplitude[0]) / AP_amplitude[0]
LibV5 : AP_amplitude_from_voltagebase
The relative height of the action potential from voltage base
Required features: LibV5:voltage_base, LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
AP_amplitude_from_voltagebase = peak_voltage - voltage_base
LibV5 : AP1_peak, AP2_peak
The peak voltage of the first and second action potentials
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
AP1_peak = peak_voltage[0] AP2_peak = peak_voltage[1]
LibV5 : AP2_AP1_diff
Difference amplitude of the second to first spike
Required features: LibV1:AP_amplitude (mV)
Units: mV
Pseudocode:
AP2_AP1_diff = AP_amplitude[1] - AP_amplitude[0]
LibV5 : AP2_AP1_peak_diff
Difference peak voltage of the second to first spike
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
AP2_AP1_diff = peak_voltage[1] - peak_voltage[0]
LibV2 : amp_drop_first_second
Difference of the amplitude of the first and the second peak
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
amp_drop_first_second = peak_voltage[0] - peak_voltage[1]
LibV2 : amp_drop_first_last
Difference of the amplitude of the first and the last peak
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
amp_drop_first_last = peak_voltage[0] - peak_voltage[-1]
LibV2 : amp_drop_second_last
Difference of the amplitude of the second and the last peak
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
amp_drop_second_last = peak_voltage[1] - peak_voltage[-1]
LibV2 : max_amp_difference
Maximum difference of the height of two subsequent peaks
Required features: LibV1:peak_voltage (mV)
Units: mV
Pseudocode:
max_amp_difference = numpy.max(peak_voltage[:-1] - peak_voltage[1:])
LibV1 : AP_amplitude_diff
Difference of the amplitude of two subsequent peaks
Required features: LibV1:AP_amplitude (mV)
Units: mV
Pseudocode:
AP_amplitude_diff = AP_amplitude[1:] - AP_amplitude[:-1]
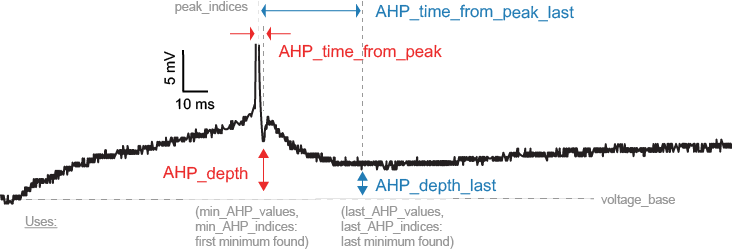
LibV5 : min_AHP_values
Absolute voltage values at the first after-hyperpolarization.
Required features: LibV5:min_AHP_indices
Units: mV
LibV5 : AHP_depth_abs
Absolute voltage values at the first after-hyperpolarization. Is the same as min_AHP_values
Required features: LibV5:min_AHP_values (mV)
Units: mV
LibV1 : AHP_depth_abs_slow
Absolute voltage values at the first after-hyperpolarization starting a given number of ms (default: 5) after the peak
Required features: LibV1:peak_indices
Units: mV
LibV1 : AHP_depth_slow
Relative voltage values at the first after-hyperpolarization starting a given number of ms (default: 5) after the peak
Required features: LibV5:voltage_base (mV), LibV1:AHP_depth_abs_slow (mV)
Units: mV
Pseudocode:
AHP_depth_slow = AHP_depth_abs_slow[:] - voltage_base
LibV1 : AHP_slow_time
Time difference between slow AHP (see AHP_depth_abs_slow) and peak, divided by interspike interval
Required features: LibV1:AHP_depth_abs_slow
Units: constant
LibV1 : AHP_depth
Relative voltage values at the first after-hyperpolarization
Required features: LibV5:voltage_base (mV), LibV5:min_AHP_values (mV)
Units: mV
Pseudocode:
min_AHP_values = first_min_element(voltage, peak_indices) AHP_depth = min_AHP_values[:] - voltage_base
LibV1 : AHP_depth_diff
Difference of subsequent relative voltage values at the first after-hyperpolarization
Required features: LibV1:AHP_depth (mV)
Units: mV
Pseudocode:
AHP_depth_diff = AHP_depth[1:] - AHP_depth[:-1]
LibV2 : fast_AHP
Voltage value of the action potential onset relative to the subsequent AHP
Ignores the last spike
Required features: LibV5:AP_begin_indices, LibV5:min_AHP_values
Units: mV
Pseudocode:
fast_AHP = voltage[AP_begin_indices[:-1]] - voltage[min_AHP_indices[:-1]]
LibV2 : fast_AHP_change
Difference of the fast AHP of the second and the first action potential divided by the fast AHP of the first action potential
Required features: LibV2:fast_AHP
Units: constant
Pseudocode:
fast_AHP_change = (fast_AHP[1:] - fast_AHP[0]) / fast_AHP[0]
LibV5 : AHP_depth_from_peak, AHP1_depth_from_peak, AHP2_depth_from_peak
Voltage difference between AP peaks and first AHP depths
Required features: LibV1:peak_indices, LibV5:min_AHP_indices
Units: mV
Pseudocode:
AHP_depth_from_peak = v[peak_indices] - v[min_AHP_indices] AHP1_depth_from_peak = AHP_depth_from_peak[0] AHP2_depth_from_peak = AHP_depth_from_peak[1]
LibV5 : AHP_time_from_peak
Time between AP peaks and first AHP depths
Required features: LibV1:peak_indices, LibV5:min_AHP_values (mV)
Units: ms
Pseudocode:
min_AHP_indices = first_min_element(voltage, peak_indices) AHP_time_from_peak = t[min_AHP_indices[:]] - t[peak_indices[i]]
LibV5 : ADP_peak_values
Absolute voltage values of the small afterdepolarization peak
strict_stiminterval should be set to True for this feature to behave as expected.
Required features: LibV5:min_AHP_indices, LibV5:min_between_peaks_indices
Units: mV
Pseudocode:
adp_peak_values = numpy.array( [numpy.max(v[i:j + 1]) for (i, j) in zip(min_AHP_indices, min_v_indices)] )
LibV5 : ADP_peak_amplitude
Amplitude of the small afterdepolarization peak with respect to the fast AHP voltage
strict_stiminterval should be set to True for this feature to behave as expected.
Required features: LibV5:min_AHP_values, LibV5:ADP_peak_values
Units: mV
Pseudocode:
adp_peak_amplitude = adp_peak_values - min_AHP_values
LibV3 : depolarized_base
Mean voltage between consecutive spikes (from the end of one spike to the beginning of the next one)
Required features: LibV5:AP_end_indices, LibV5:AP_begin_indices
Units: mV
Pseudocode:
depolarized_base = [] for (start_idx, end_idx) in zip( AP_end_indices[:-1], AP_begin_indices[1:]) ): depolarized_base.append(numpy.mean(voltage[start_idx:end_idx]))
LibV5 : min_voltage_between_spikes
Minimal voltage between consecutive spikes
Required features: LibV5:peak_indices
Units: mV
Pseudocode:
min_voltage_between_spikes = [] for peak1, peak2 in zip(peak_indices[:-1], peak_indices[1:]): min_voltage_between_spikes.append(numpy.min(voltage[peak1:peak2]))
LibV5 : min_between_peaks_values
Minimal voltage between consecutive spikes
The last value of min_between_peaks_values is the minimum between last spike and stimulus end if strict stiminterval is True, and minimum between last spike and last voltage value if strict stiminterval is False
Required features: LibV5:min_between_peaks_indices
Units: mV
Pseudocode:
min_between_peaks_values = v[min_between_peaks_indices]
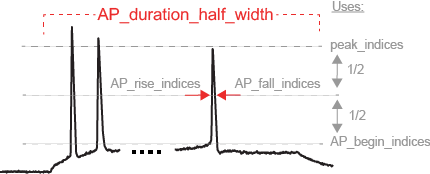
LibV2 : AP_duration_half_width
Width of spike at half spike amplitude, with spike onset as described in LibV5: AP_begin_time
Required features: LibV2: AP_rise_indices, LibV2: AP_fall_indices
Units: ms
Pseudocode:
AP_rise_indices = index_before_peak((v(peak_indices) - v(AP_begin_indices)) / 2) AP_fall_indices = index_after_peak((v(peak_indices) - v(AP_begin_indices)) / 2) AP_duration_half_width = t(AP_fall_indices) - t(AP_rise_indices)
LibV2 : AP_duration_half_width_change
Difference of the FWHM of the second and the first action potential divided by the FWHM of the first action potential
Required features: LibV2: AP_duration_half_width
Units: constant
Pseudocode:
AP_duration_half_width_change = ( AP_duration_half_width[1:] - AP_duration_half_width[0] ) / AP_duration_half_width[0]
LibV1 : AP_width
Width of spike at threshold, bounded by minimum AHP
Can use strict_stiminterval compute only for data in stimulus interval.
Required features: LibV1: peak_indices, LibV5: min_AHP_indices, threshold
Units: ms
Pseudocode:
min_AHP_indices = numpy.concatenate([[stim_start], min_AHP_indices]) for i in range(len(min_AHP_indices)-1): onset_index = numpy.where(v[min_AHP_indices[i]:min_AHP_indices[i+1]] > threshold)[0] onset_time[i] = t[onset_index] offset_time[i] = t[numpy.where(v[onset_index:min_AHP_indices[i+1]] < threshold)[0]] AP_width[i] = t(offset_time[i]) - t(onset_time[i])
LibV2 : AP_duration
Duration of an action potential from onset to offset
Required features: LibV5:AP_begin_indices, LibV5:AP_end_indices
Units: ms
Pseudocode:
AP_duration = time[AP_end_indices] - time[AP_begin_indices]
LibV2 : AP_duration_change
Difference of the durations of the second and the first action potential divided by the duration of the first action potential
Required features: LibV2:AP_duration
Units: constant
Pseudocode:
AP_duration_change = (AP_duration[1:] - AP_duration[0]) / AP_duration[0]
LibV5 : AP_width_between_threshold
Width of spike at threshold, bounded by minimum between peaks
Can use strict_stiminterval to not use minimum after stimulus end.
Required features: LibV1: peak_indices, LibV5: min_between_peaks_indices, threshold
Units: ms
Pseudocode:
min_between_peaks_indices = numpy.concatenate([[stim_start], min_between_peaks_indices]) for i in range(len(min_between_peaks_indices)-1): onset_index = numpy.where(v[min_between_peaks_indices[i]:min_between_peaks_indices[i+1]] > threshold)[0] onset_time[i] = t[onset_index] offset_time[i] = t[numpy.where(v[onset_index:min_between_peaks_indices[i+1]] < threshold)[0]] AP_width[i] = t(offset_time[i]) - t(onset_time[i])
LibV5 : spike_half_width, AP1_width, AP2_width, APlast_width
Width of spike at half spike amplitude, with the spike amplitude taken as the difference between the minimum between two peaks and the next peak
Required features: LibV5: peak_indices, LibV5: min_AHP_indices
Units: ms
Pseudocode:
min_AHP_indices = numpy.concatenate([[stim_start], min_AHP_indices]) for i in range(1, len(min_AHP_indices)): v_half_width = (v[peak_indices[i-1]] + v[min_AHP_indices[i]]) / 2. rise_idx = numpy.where(v[min_AHP_indices[i-1]:peak_indices[i-1]] > v_half_width)[0] v_dev = v_half_width - v[rise_idx] delta_v = v[rise_idx] - v[rise_idx - 1] delta_t = t[rise_idx] - t[rise_idx - 1] t_dev_rise = delta_t * v_dev / delta_v fall_idx = numpy.where(v[peak_indices[i-1]:min_AHP_indices[i]] < v_half_width)[0] v_dev = v_half_width - v[fall_idx] delta_v = v[fall_idx] - v[fall_idx - 1] delta_t = t[fall_idx] - t[fall_idx - 1] t_dev_fall = delta_t * v_dev / delta_v spike_half_width[i] = t[fall_idx] + t_dev_fall - t[rise_idx] - t_dev_rise AP1_width = spike_half_width[0] AP2_width = spike_half_width[1] APlast_width = spike_half_width[-1]
LibV1 : spike_width2
Width of spike at half spike amplitude, with the spike onset taken as the maximum of the second derivative of the voltage in the range between the minimum between two peaks and the next peak
Required features: LibV5: peak_indices, LibV5: min_AHP_indices
Units: ms
Pseudocode:
for i in range(len(min_AHP_indices)): dv2 = CentralDiffDerivative(CentralDiffDerivative(v[min_AHP_indices[i]:peak_indices[i + 1]])) peak_onset_idx = numpy.argmax(dv2) + min_AHP_indices[i] v_half_width = (v[peak_indices[i + 1]] + v[peak_onset_idx]) / 2. rise_idx = numpy.where(v[peak_onset_idx:peak_indices[i + 1]] > v_half_width)[0] v_dev = v_half_width - v[rise_idx] delta_v = v[rise_idx] - v[rise_idx - 1] delta_t = t[rise_idx] - t[rise_idx - 1] t_dev_rise = delta_t * v_dev / delta_v fall_idx = numpy.where(v[peak_indices[i + 1]:] < v_half_width)[0] v_dev = v_half_width - v[fall_idx] delta_v = v[fall_idx] - v[fall_idx - 1] delta_t = t[fall_idx] - t[fall_idx - 1] t_dev_fall = delta_t * v_dev / delta_v spike_width2[i] = t[fall_idx] + t_dev_fall - t[rise_idx] - t_dev_rise
LibV5 : AP_begin_width, AP1_begin_width, AP2_begin_width
Width of spike at spike start
Required features: LibV5: min_AHP_indices, LibV5: AP_begin_indices
Units: ms
Pseudocode:
for i in range(len(min_AHP_indices)): rise_idx = AP_begin_indices[i] fall_idx = numpy.where(v[rise_idx + 1:min_AHP_indices[i]] < v[rise_idx])[0] AP_begin_width[i] = t[fall_idx] - t[rise_idx] AP1_begin_width = AP_begin_width[0] AP2_begin_width = AP_begin_width[1]
LibV5 : AP2_AP1_begin_width_diff
Difference width of the second to first spike
Required features: LibV5: AP_begin_width
Units: ms
Pseudocode:
AP2_AP1_begin_width_diff = AP_begin_width[1] - AP_begin_width[0]
LibV5 : AP_begin_voltage, AP1_begin_voltage, AP2_begin_voltage
Voltage at spike start
Required features: LibV5: AP_begin_indices
Units: mV
Pseudocode:
AP_begin_voltage = v[AP_begin_indices] AP1_begin_voltage = AP_begin_voltage[0] AP2_begin_voltage = AP_begin_voltage[1]
LibV5 : AP_begin_time
Time at spike start. Spike start is defined as where the first derivative of the voltage trace is higher than 10 V/s , for at least 5 points
Required features: LibV5: AP_begin_indices
Units: ms
Pseudocode:
AP_begin_time = t[AP_begin_indices]
LibV5 : AP_peak_upstroke
Maximum of rise rate of spike
Required features: LibV5: AP_begin_indices, LibV5: peak_indices
Units: V/s
Pseudocode:
ap_peak_upstroke = [] for apbi, pi in zip(ap_begin_indices, peak_indices): ap_peak_upstroke.append(numpy.max(dvdt[apbi:pi]))
LibV5 : AP_peak_downstroke
Minimum of fall rate from spike
Required features: LibV5: min_AHP_indices, LibV5: peak_indices
Units: V/s
Pseudocode:
ap_peak_downstroke = [] for ahpi, pi in zip(min_ahp_indices, peak_indices): ap_peak_downstroke.append(numpy.min(dvdt[pi:ahpi]))
LibV2 : AP_rise_time
Time between the AP threshold and the peak, given a window (default: from 0% to 100% of the AP amplitude)
Required features: LibV5: AP_begin_indices, LibV5: peak_indices, LibV1: AP_amplitude
Units: ms
Pseudocode:
rise_times = [] begin_voltages = AP_amps * rise_start_perc + voltage[AP_begin_indices] end_voltages = AP_amps * rise_end_perc + voltage[AP_begin_indices] for AP_begin_indice, peak_indice, begin_v, end_v in zip( AP_begin_indices, peak_indices, begin_voltages, end_voltages ): voltage_window = voltage[AP_begin_indice:peak_indice] new_begin_indice = AP_begin_indice + numpy.min( numpy.where(voltage_window >= begin_v)[0] ) new_end_indice = AP_begin_indice + numpy.max( numpy.where(voltage_window <= end_v)[0] ) rise_times.append(time[new_end_indice] - time[new_begin_indice])
LibV2 : AP_fall_time
Time from action potential maximum to the offset
Required features: LibV5: AP_end_indices, LibV5: peak_indices
Units: ms
Pseudocode:
AP_fall_time = time[AP_end_indices] - time[peak_indices]
LibV2 : AP_rise_rate
Voltage change rate during the rising phase of the action potential
Required features: LibV5: AP_begin_indices, LibV5: peak_indices
Units: V/s
Pseudocode:
AP_rise_rate = (voltage[peak_indices] - voltage[AP_begin_indices]) / ( time[peak_indices] - time[AP_begin_indices] )
LibV2 : AP_fall_rate
Voltage change rate during the falling phase of the action potential
Required features: LibV5: AP_end_indices, LibV5: peak_indices
Units: V/s
Pseudocode:
AP_fall_rate = (voltage[AP_end_indices] - voltage[peak_indices]) / ( time[AP_end_indices] - time[peak_indices] )
LibV2 : AP_rise_rate_change
Difference of the rise rates of the second and the first action potential divided by the rise rate of the first action potential
Required features: LibV2: AP_rise_rate_change
Units: constant
Pseudocode:
AP_rise_rate_change = (AP_rise_rate[1:] - AP_rise_rate[0]) / AP_rise_rate[0]
LibV2 : AP_fall_rate_change
Difference of the fall rates of the second and the first action potential divided by the fall rate of the first action potential
Required features: LibV2: AP_fall_rate_change
Units: constant
Pseudocode:
AP_fall_rate_change = (AP_fall_rate[1:] - AP_fall_rate[0]) / AP_fall_rate[0]
LibV5 : AP_phaseslope
Slope of the V, dVdt phasespace plot at the beginning of every spike
(at the point where the derivative crosses the DerivativeThreshold)
Required features: LibV5:AP_begin_indices
Parameters: AP_phaseslope_range
Units: 1/(ms)
Pseudocode:
range_max_idxs = AP_begin_indices + AP_phseslope_range range_min_idxs = AP_begin_indices - AP_phseslope_range AP_phaseslope = (dvdt[range_max_idxs] - dvdt[range_min_idxs]) / (v[range_max_idxs] - v[range_min_idxs])
Voltage features

LibV5 : steady_state_voltage_stimend
The average voltage during the last 10% of the stimulus duration.
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
stim_duration = stim_end - stim_start begin_time = stim_end - 0.1 * stim_duration end_time = stim_end steady_state_voltage_stimend = numpy.mean(voltage[numpy.where((t < end_time) & (t >= begin_time))])
LibV2 : steady_state_hyper
Steady state voltage during hyperpolarization for 30 data points (after interpolation)
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
stim_end_idx = numpy.argwhere(time >= stim_end)[0][0] steady_state_hyper = numpy.mean(voltage[stim_end_idx - 35:stim_end_idx - 5])
LibV1 : steady_state_voltage
The average voltage after the stimulus
Required features: t, V, stim_end
Units: mV
Pseudocode:
steady_state_voltage = numpy.mean(voltage[numpy.where((t <= max(t)) & (t > stim_end))])
LibV5 : voltage_base
The average voltage during the last 10% of time before the stimulus.
Required features: t, V, stim_start, stim_end
Parameters: voltage_base_start_perc (default = 0.9), voltage_base_end_perc (default = 1.0)
Units: mV
Pseudocode:
voltage_base = numpy.mean(voltage[numpy.where( (t >= voltage_base_start_perc * stim_start) & (t <= voltage_base_end_perc * stim_start))])
LibV5 : current_base
The average current during the last 10% of time before the stimulus.
Required features: t, I, stim_start, stim_end
Parameters: current_base_start_perc (default = 0.9), current_base_end_perc (default = 1.0), precision_threshold (default = 1e-10), current_base_mode (can be “mean” or “median”, default=”mean”)
Units: nA
Pseudocode:
current_slice = I[numpy.where( (t >= current_base_start_perc * stim_start) & (t <= current_base_end_perc * stim_start))] if current_base_mode == "mean": current_base = numpy.mean(current_slice) elif current_base_mode == "median": current_base = numpy.median(current_slice)
LibV1 : time_constant
The membrane time constant
The extraction of the time constant requires a voltage trace of a cell in a hyper- polarized state. Starting at stim start find the beginning of the exponential decay where the first derivative of V(t) is smaller than -0.005 V/s in 5 subsequent points. The flat subsequent to the exponential decay is defined as the point where the first derivative of the voltage trace is bigger than -0.005 and the mean of the follwowing 70 points as well. If the voltage trace between the beginning of the decay and the flat includes more than 9 points, fit an exponential decay. Yield the time constant of that decay.
Required features: t, V, stim_start, stim_end
Units: ms
Pseudocode:
min_derivative = 5e-3 decay_start_min_length = 5 # number of indices min_length = 10 # number of indices t_length = 70 # in ms # get start and middle indices stim_start_idx = numpy.where(time >= stim_start)[0][0] # increment stimstartindex to skip a possible transient stim_start_idx += 10 stim_middle_idx = numpy.where(time >= (stim_start + stim_end) / 2.)[0][0] # get derivative t_interval = time[stim_start_idx:stim_middle_idx] dv = five_point_stencil_derivative(voltage[stim_start_idx:stim_middle_idx]) dt = five_point_stencil_derivative(t_interval) dvdt = dv / dt # find start and end of decay # has to be over deriv threshold for at least a given number of indices pass_threshold_idxs = numpy.append( -1, numpy.argwhere(dvdt > -min_derivative).flatten() ) length_idx = numpy.argwhere( numpy.diff(pass_threshold_idxs) > decay_start_min_length )[0][0] i_start = pass_threshold_idxs[length_idx] + 1 # find flat (end of decay) flat_idxs = numpy.argwhere(dvdt[i_start:] > -min_derivative).flatten() # for loop is not optimised # but we expect the 1st few values to be the ones we are looking for for i in flat_idxs: i_flat = i + i_start i_flat_stop = numpy.argwhere( t_interval >= t_interval[i_flat] + t_length )[0][0] if numpy.mean(dvdt[i_flat:i_flat_stop]) > -min_derivative: break dvdt_decay = dvdt[i_start:i_flat] t_decay = time[stim_start_idx + i_start:stim_start_idx + i_flat] v_decay_tmp = voltage[stim_start_idx + i_start:stim_start_idx + i_flat] v_decay = abs(v_decay_tmp - voltage[stim_start_idx + i_flat]) if len(dvdt_decay) < min_length: return None # -- golden search algorithm -- # from scipy.optimize import minimize_scalar def numpy_fit(x, t_decay, v_decay): new_v_decay = v_decay + x log_v_decay = numpy.log(new_v_decay) (slope, _), res, _, _, _ = numpy.polyfit( t_decay, log_v_decay, 1, full=True ) range = numpy.max(log_v_decay) - numpy.min(log_v_decay) return res / (range * range) max_bound = min_derivative * 1000. golden_bracket = [0, max_bound] result = minimize_scalar( numpy_fit, args=(t_decay, v_decay), bracket=golden_bracket, method='golden', ) # -- fit -- # log_v_decay = numpy.log(v_decay + result.x) slope, _ = numpy.polyfit(t_decay, log_v_decay, 1) time_constant = -1. / slope
LibV5 : decay_time_constant_after_stim
The decay time constant of the voltage right after the stimulus
Required features: t, V, stim_start, stim_end
Parameters: decay_start_after_stim (default = 1.0 ms), decay_end_after_stim (default = 10.0 ms)
Units: ms
Pseudocode:
time_interval = t[numpy.where(t => decay_start_after_stim & t < decay_end_after_stim)] - t[numpy.where(t == stim_end)] voltage_interval = abs(voltages[numpy.where(t => decay_start_after_stim & t < decay_end_after_stim)] - voltages[numpy.where(t == decay_start_after_stim)]) log_voltage_interval = numpy.log(voltage_interval) slope, _ = numpy.polyfit(time_interval, log_voltage_interval, 1) decay_time_constant_after_stim = -1. / slope
LibV5 : multiple_decay_time_constant_after_stim
When multiple stimuli are applied, this function returns a list of decay time constants each computed on the voltage right after each stimulus.
The settings multi_stim_start and multi_stim_end are mandatory for this feature to work. Each is a list containing the start and end times of each stimulus present in the current protocol respectively.
Required features: t, V, stim_start, stim_end
Required settings: multi_stim_start, multi_stim_end
Parameters: decay_start_after_stim (default = 1.0 ms), decay_end_after_stim (default = 10.0 ms)
Units: ms
Pseudocode:
multiple_decay_time_constant_after_stim = [] for i in range(len(number_stimuli): stim_start = multi_stim_start[i] stim_end = multi_stim_end[i] multiple_decay_time_constant_after_stim.append( decay_time_constant_after_stim(stim_start, stim_end) )
LibV5 : sag_time_constant
The decay time constant of the exponential voltage decay from the bottom of the sag to the steady-state.
The start of the decay is taken at the minimum voltage (the bottom of the sag). The end of the decay is taken when the voltage crosses the steady state voltage minus 10% of the sag amplitude. The time constant is the slope of the linear fit to the log of the voltage. The golden search algorithm is not used, since the data is expected to be noisy and adding a parameter in the log ( log(voltage + x) ) is likely to increase errors on the fit.
Required features: t, V, stim_start, stim_end, minimum_voltage, steady_state_voltage_stimend, sag_amplitude
Units: ms
Pseudocode:
# get start decay start_decay = numpy.argmin(vinterval) # get end decay v90 = steady_state_v - 0.1 * sag_ampl end_decay = numpy.where((tinterval > tinterval[start_decay]) & (vinterval >= v90))[0][0] v_reference = vinterval[end_decay] # select t, v in decay interval interval_indices = numpy.arange(start_decay, end_decay) interval_time = tinterval[interval_indices] interval_voltage = abs(vinterval[interval_indices] - v_reference) # get tau log_interval_voltage = numpy.log(interval_voltage) slope, _ = numpy.polyfit(interval_time, log_interval_voltage, 1) tau = abs(1. / slope)
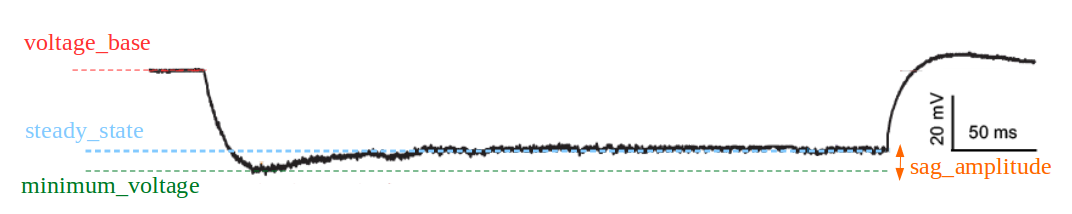
LibV5 : sag_amplitude
The difference between the minimal voltage and the steady state at stimend
Required features: t, V, stim_start, stim_end, steady_state_voltage_stimend, minimum_voltage, voltage_deflection_stim_ssse
Parameters:
Units: mV
Pseudocode:
if (voltage_deflection_stim_ssse <= 0): sag_amplitude = steady_state_voltage_stimend - minimum_voltage else: sag_amplitude = None
LibV5 : sag_ratio1
The ratio between the sag amplitude and the maximal sag extend from voltage base
Required features: t, V, stim_start, stim_end, sag_amplitude, voltage_base, minimum_voltage
Parameters:
Units: constant
Pseudocode:
if voltage_base != minimum_voltage: sag_ratio1 = sag_amplitude / (voltage_base - minimum_voltage) else: sag_ratio1 = None
LibV5 : sag_ratio2
The ratio between the maximal extends of sag from steady state and voltage base
Required features: t, V, stim_start, stim_end, steady_state_voltage_stimend, voltage_base, minimum_voltage
Parameters:
Units: constant
Pseudocode:
if voltage_base != minimum_voltage: sag_ratio2 = (voltage_base - steady_state_voltage_stimend) / (voltage_base - minimum_voltage) else: sag_ratio2 = None
LibV1 : ohmic_input_resistance
The ratio between the voltage deflection and stimulus current
Required features: t, V, stim_start, stim_end, voltage_deflection
Parameters: stimulus_current
Units: MΩ
Pseudocode:
ohmic_input_resistance = voltage_deflection / stimulus_current
LibV5 : ohmic_input_resistance_vb_ssse
The ratio between the voltage deflection (between voltage base and steady-state voltage at stimend) and stimulus current
Required features: t, V, stim_start, stim_end, voltage_deflection_vb_ssse
Parameters: stimulus_current
Units: MΩ
Pseudocode:
ohmic_input_resistance_vb_ssse = voltage_deflection_vb_ssse / stimulus_current
LibV5 : voltage_deflection_vb_ssse
The voltage deflection between voltage base and steady-state voltage at stimend
The voltage base used is the average voltage during the last 10% of time before the stimulus and the steady state voltage at stimend used is the average voltage during the last 10% of the stimulus duration.
Required features: t, V, stim_start, stim_end, voltage_base, steady_state_voltage_stimend
Units: mV
Pseudocode:
voltage_deflection_vb_ssse = steady_state_voltage_stimend - voltage_base
LibV1 : voltage_deflection
The voltage deflection between voltage base and steady-state voltage at stimend
The voltage base used is the average voltage during all of the time before the stimulus and the steady state voltage at stimend used is the average voltage of the five values before the last five values before the end of the stimulus duration.
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
voltage_base = numpy.mean(V[t < stim_start]) stim_end_idx = numpy.where(t >= stim_end)[0][0] steady_state_voltage_stimend = numpy.mean(V[stim_end_idx-10:stim_end_idx-5]) voltage_deflection = steady_state_voltage_stimend - voltage_base
LibV5 : voltage_deflection_begin
The voltage deflection between voltage base and steady-state voltage soon after stimulation start
The voltage base used is the average voltage during all of the time before the stimulus and the steady state voltage used is the average voltage taken from 5% to 15% of the stimulus duration.
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
voltage_base = numpy.mean(V[t < stim_start]) tstart = stim_start + 0.05 * (stim_end - stim_start) tend = stim_start + 0.15 * (stim_end - stim_start) condition = numpy.all((tstart < t, t < tend), axis=0) steady_state_voltage_stimend = numpy.mean(V[condition]) voltage_deflection = steady_state_voltage_stimend - voltage_base
LibV5 : voltage_after_stim
The mean voltage after the stimulus in (stim_end + 25%*end_period, stim_end + 75%*end_period)
Required features: t, V, stim_end
Units: mV
Pseudocode:
tstart = stim_end + (t[-1] - stimEnd) * 0.25 tend = stim_end + (t[-1] - stimEnd) * 0.75 condition = numpy.all((tstart < t, t < tend), axis=0) voltage_after_stim = numpy.mean(V[condition])
LibV1 : minimum_voltage
The minimum of the voltage during the stimulus
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
minimum_voltage = min(voltage[numpy.where((t >= stim_start) & (t <= stim_end))])
LibV1 : maximum_voltage
The maximum of the voltage during the stimulus
Required features: t, V, stim_start, stim_end
Units: mV
Pseudocode:
maximum_voltage = max(voltage[numpy.where((t >= stim_start) & (t <= stim_end))])
LibV5 : maximum_voltage_from_voltagebase
Difference between maximum voltage during stimulus and voltage base
Required features: maximum_voltage, voltage_base
Units: mV
Pseudocode:
maximum_voltage_from_voltagebase = maximum_voltage - voltage_base
Requested eFeatures
Cpp features
LibV1 : AHP_depth_last
Relative voltage values at the last after-hyperpolarization
Required features: LibV5:voltage_base (mV), LibV5:last_AHP_values (mV)
Units: mV
Pseudocode:
last_AHP_values = last_min_element(voltage, peak_indices) AHP_depth = last_AHP_values[:] - voltage_base
LibV5 : AHP_time_from_peak_last
Time between AP peaks and last AHP depths
Required features: LibV1:peak_indices, LibV5:min_AHP_values (mV)
Units: ms
Pseudocode:
last_AHP_indices = last_min_element(voltage, peak_indices) AHP_time_from_peak_last = t[last_AHP_indices[:]] - t[peak_indices[i]]
LibV5 : steady_state_voltage_stimend_from_voltage_base
The average voltage during the last 90% of the stimulus duration realtive to voltage_base
Required features: LibV5: steady_state_voltage_stimend (mV), LibV5: voltage_base (mV)
Units: mV
Pseudocode:
steady_state_voltage_stimend_from_voltage_base = steady_state_voltage_stimend - voltage_base
LibV5 : min_duringstim_from_voltage_base
The minimum voltage during stimulus
Required features: LibV5: min_duringstim (mV), LibV5: voltage_base (mV)
Units: mV
Pseudocode:
min_duringstim_from_voltage_base = minimum_voltage - voltage_base
LibV5 : max_duringstim_from_voltage_base
The minimum voltage during stimulus
Required features: LibV5: max_duringstim (mV), LibV5: voltage_base (mV)
Units: mV
Pseudocode:
max_duringstim_from_voltage_base = maximum_voltage - voltage_base
LibV5 : diff_max_duringstim
Difference between maximum and steady state during stimulation
Required features: LibV5: max_duringstim (mV), LibV5: steady_state_voltage_stimend (mV)
Units: mV
Pseudocode:
diff_max_duringstim: max_duringstim - steady_state_voltage_stimend
LibV5 : diff_min_duringstim
Difference between minimum and steady state during stimulation
Required features: LibV5: min_duringstim (mV), LibV5: steady_state_voltage_stimend (mV)
Units: mV
Pseudocode:
diff_min_duringstim: min_duringstim - steady_state_voltage_stimend
Python features
Python efeature : initburst_sahp
Slow AHP voltage after initial burst
The end of the initial burst is detected when the ISIs frequency gets lower than initburst_freq_threshold, in Hz. Then the sahp is searched for the interval between initburst_sahp_start (in ms) after the last spike of the burst, and initburst_sahp_end (in ms) after the last spike of the burst.
Required features: LibV1: peak_time
Parameters: initburst_freq_threshold (default=50), initburst_sahp_start (default=5), initburst_sahp_end (default=100)
Units: mV
Python efeature : initburst_sahp_ssse
Slow AHP voltage from steady_state_voltage_stimend after initial burst
Required features: LibV5: steady_state_voltage_stimend, initburst_sahp
Units: mV
Pseudocode:
numpy.array([initburst_sahp_value[0] - ssse[0]])
Python efeature : initburst_sahp_vb
Slow AHP voltage from voltage base after initial burst
Required features: LibV5: voltage_base, initburst_sahp
Units: mV
Pseudocode:
numpy.array([initburst_sahp_value[0] - voltage_base[0]])
Python efeature : depol_block_bool
Check for a depolarization block. Returns 1 if there is a depolarization block or a hyperpolarization block, and returns 0 otherwise.
A depolarization block is detected when the voltage stays higher than the mean of AP_begin_voltage for longer than 50 ms.
A hyperpolarization block is detected when, after stimulus start, the voltage stays below -75 mV for longer than 50 ms.
Required features: LibV5: AP_begin_voltage
Units: constant
Python efeature : spikes_per_burst
Number of spikes in each burst.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Defalt value is 2.0.
Required features: LibV5: burst_begin_indices, LibV5: burst_end_indices
Units: constant
Pseudocode:
spike_per_bursts = [] for idx_begin, idx_end in zip(burst_begin_indices, burst_end_indices): spike_per_bursts.append(idx_end - idx_begin + 1)
Python efeature : spikes_per_burst_diff
Difference of number of spikes between each burst and the next one.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Defalt value is 2.0.
Required features: spikes_per_burst
Units: constant
Pseudocode:
spikes_per_burst[:-1] - spikes_per_burst[1:]
Python efeature : spikes_in_burst1_burst2_diff
Difference of number of spikes between the first burst and the second one.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Defalt value is 2.0.
Required features: spikes_per_burst_diff
Units: constant
Pseudocode:
numpy.array([spikes_per_burst_diff[0]])
Python efeature : spikes_in_burst1_burstlast_diff
Difference of number of spikes between the first burst and the last one.
The first spike is ignored by default. This can be changed by setting ignore_first_ISI to 0.
The burst detection can be fine-tuned by changing the setting strict_burst_factor. Defalt value is 2.0.
Required features: spikes_per_burst
Units: constant
Pseudocode:
numpy.array([spikes_per_burst[0] - spikes_per_burst[-1]])
Python efeature : impedance
Computes the impedance given a ZAP current input and its voltage response. It will return the frequency at which the impedance is maximal, in the range (0, impedance_max_freq] Hz, with impedance_max_freq being a setting with 50.0 as a default value.
Required features: current, spike_count, LibV5:voltage_base, LibV5:current_base
Units: Hz
Pseudocode:
normalized_voltage = voltage_trace - voltage_base normalized_current = current_trace - current_base if spike_count < 1: # if there is no spikes in ZAP fft_volt = numpy.fft.fft(normalized_voltage) fft_cur = numpy.fft.fft(normalized_current) if any(fft_cur) == 0: return None # convert dt from ms to s to have freq in Hz freq = numpy.fft.fftfreq(len(normalized_voltage), d=dt / 1000.) Z = fft_volt / fft_cur norm_Z = abs(Z) / max(abs(Z)) select_idxs = numpy.swapaxes(numpy.argwhere((freq > 0) & (freq <= impedance_max_freq)), 0, 1)[0] smooth_Z = gaussian_filter1d(norm_Z[select_idxs], 10) ind_max = numpy.argmax(smooth_Z) return freq[ind_max] else: return None
Python efeature : phaseslope_max
Computes the maximum of the phase slope. Attention, this feature is sensitive to interpolation timestep.
Required features: time, voltage
Units: V/s
Pseudocode:
phaseslope = numpy.diff(voltage) / numpy.diff(time) phaseslope_max = numpy.array([numpy.max(phaseslope)])